
Education
Date |
Affiliation |
Major |
Degree |
1999.09-2003.07 |
School of Life Sciences, Shandong University |
Biotechnology |
Bachelor |
2003.09-2008.07 |
Shanghai Institute of Materia Medica, Chinese Academy of Sciences |
Drug Design |
PhD |
Research Experience
Date |
Affiliation |
Position |
2008.08-2011.07 |
University of California, Davis, USA |
Postdoctoral Fellow |
2018.08-2016.08 |
Sanford Burnham Prebys Medical Discovery Institute at Lake Nona, USA |
Postdoctoral Associate |
2016.09- now |
State Key Laboratory of Microbial Technology, Shandong University |
Professor |
Research Interests
Our research is dedicated to investigating drug targets associated with major human diseases. Utilizing advanced techniques in structural biology and molecular pharmacology, we explore the structure and function of target proteins, elucidate protein-ligand interactions, and decipher the molecular mechanisms by which ligands modulate protein activity. In parallel, we are engaged in the discovery of novel small-molecule therapeutics through the design and implementation of diverse compound screening platforms. Lead compounds exhibiting promising activity are further optimized via computational modeling and structure-based chemical modification.
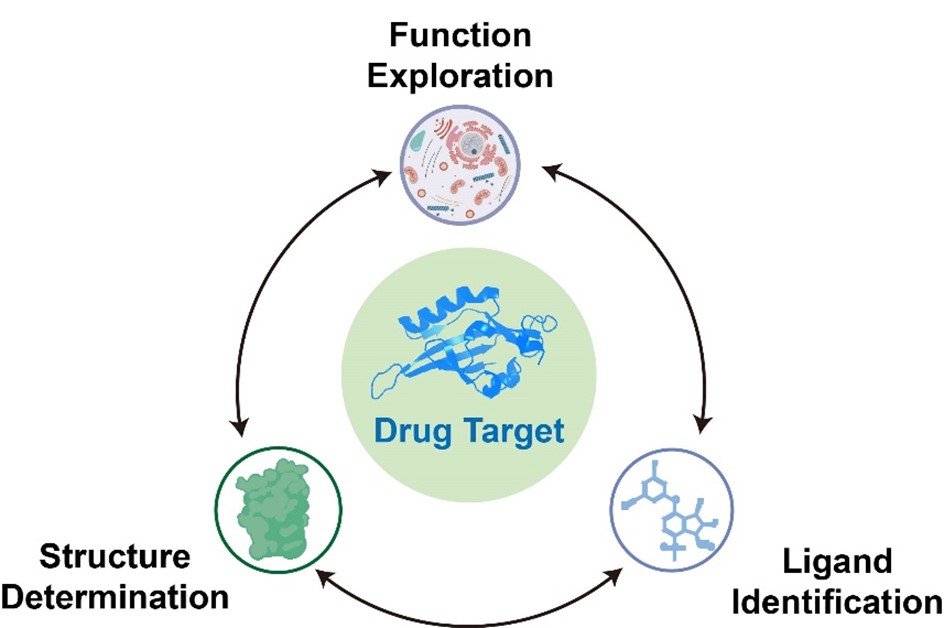
Additionally, in the field of synthetic biology, we collaborate with multiple research groups to study key enzymes within microbial biosynthetic pathways. By determining high-resolution structures of these enzymes, particularly the enzyme–substrate complexes, we aim to unravel their catalytic mechanisms and substrate specificity. These insights facilitate the rational engineering of enzymes for biotechnological and pharmaceutical applications.
Research Projects
A major focus of our current research involves “nuclear receptor-like” transcription factors, i.e. the basic helix-loop-helix-PER-ARNT-SIM (bHLH-PAS) family. These proteins are implicated in numerous human diseases and typically contain ligand-binding pockets, rendering them a promising and underexplored class of druggable transcription factors, second only to nuclear receptors.
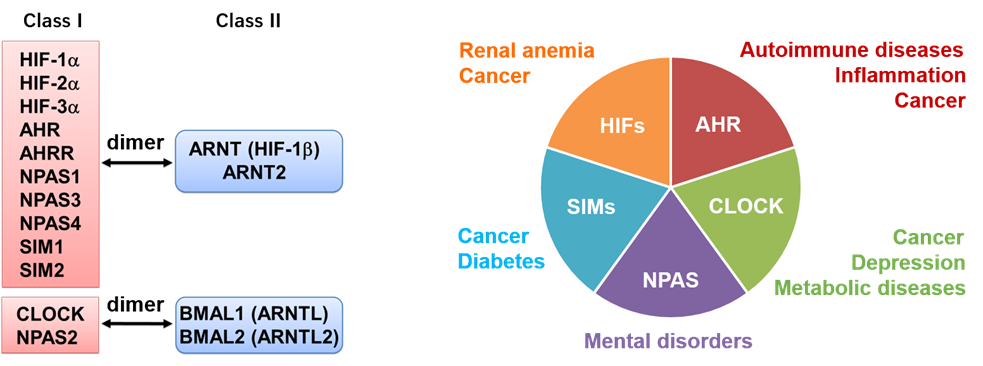
Members of the bHLH-PAS family and associated diseases
1. Bidirectional Regulation of HIF-2α Activity by Agonists and Antagonists
Hypoxia-inducible factors (HIFs), members of the bHLH-PAS family, function as oxygen sensors that orchestrate transcriptional responses to hypoxia, regulating genes involved in erythropoiesis, angiogenesis, and metabolism. HIFs operate as obligate heterodimers composed of an α subunit and a β subunit (also called ARNT). Our work has resolved complex structures of HIF-α-ARNT dimers, revealing their mode of dimerization, DNA recognition, and ligand binding. Notably, we identified the first small-molecule agonists of HIF-2α and elucidated the allosteric mechanism underlying bidirectional modulation of its transcriptional activity. These findings provide a foundation for designing new anticancer drugs (antagonists) or therapeutics for anemia (agonists) that directly target HIF-2α.
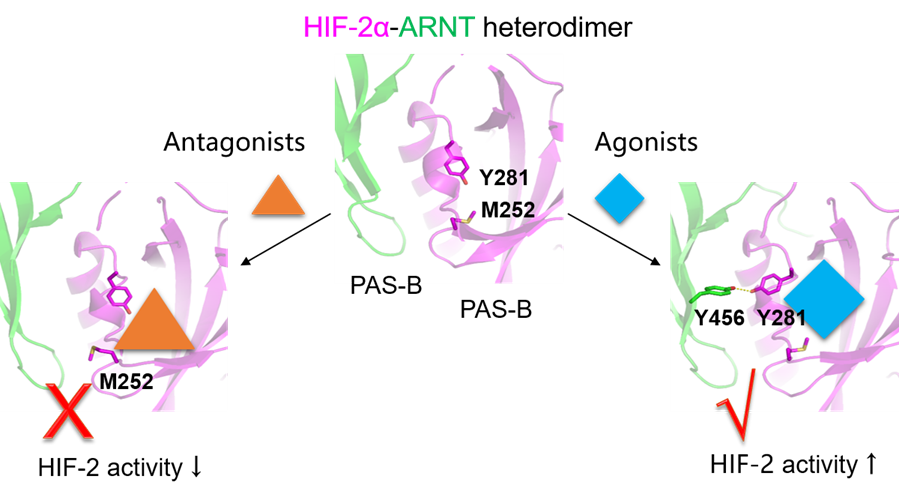
Bidirectional regulation of HIF-2α by agonists and antagonists
2. Mechanism of AHR Ligand Promiscuity and Activation Regulation
The aryl hydrocarbon receptor (AHR) exhibits remarkable ligand promiscuity, enabling it to respond to diverse molecules ranging from polycyclic aromatic hydrocarbons to endogenous metabolites. AHR plays a critical role in inflammatory diseases, autoimmune disorders, and tumor development. Understanding the structural basis of AHR ligand binding and activation is essential for developing new treatments for AHR-related pathologies. Our studies have uncovered the structural determinants of AHR recognition and binding to ligands such as Tapinarof, an anti-psoriasis drug. Furthermore, we have delineated the conformational changes and allosteric events that drive AHR activation upon ligand binding.
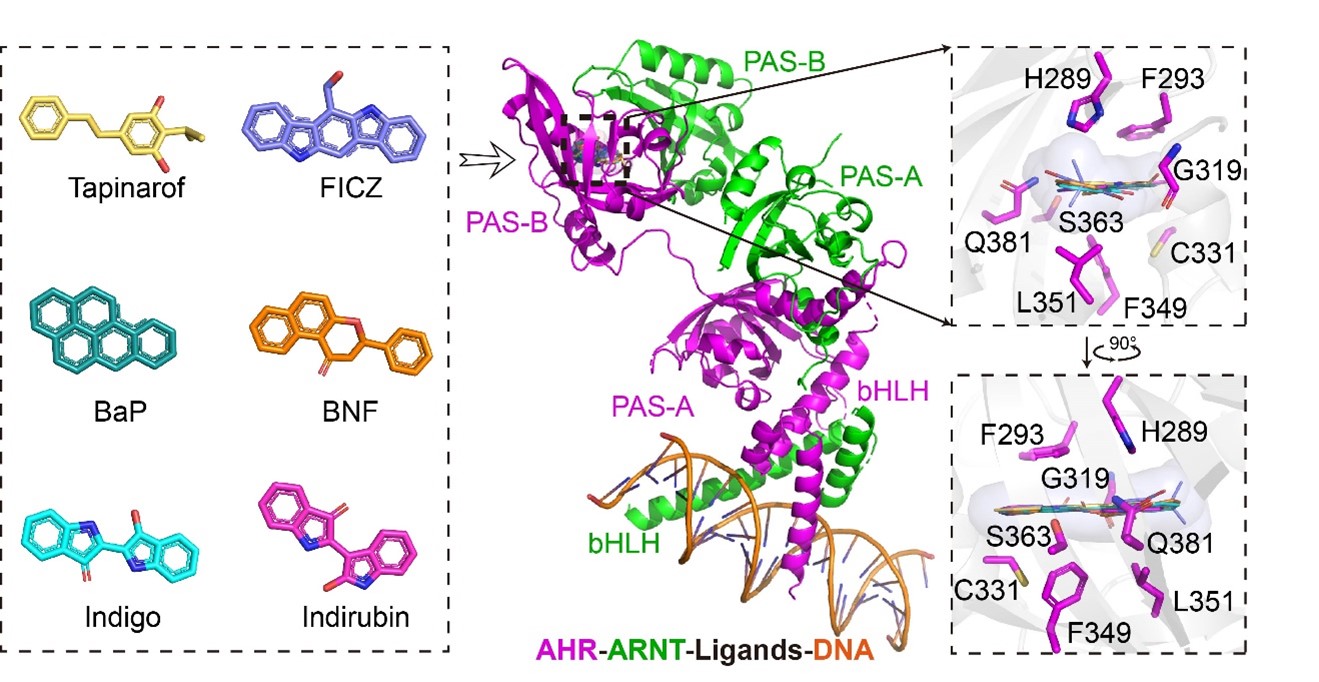
Analysis of the co-crystal structures of ligand-bound AHR
Representative Publications
A. Structural & functional studies of transcription factors and the discovery of novel ligands
1. Zhang M, Guo Y, Diao X, Guo M, Teng H, Sun X, Zhuang J, Song C, Xie X, Wu D. Molecular mechanism of the crosstalk between glucocorticoid receptor (GR) and hypoxia-inducible factor 3α (HIF-3α) pathways. Mar Life Sci Technol. 2025, Jun 4. (Corresponding)
2. Diao X, Shang Q, Guo M, Huang Y, Zhang M, Chen X, Liang Y, Sun X, Zhou F, Zhuang J, Liu SJ, Vogel CFA, Rastinejad F, Wu D. Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex. Nat Commun. 2025, 16:1282. (Corresponding Author)
3. Zhuang J, Shang Q, Rastinejad F*, Wu D*. Decoding Allosteric Control in Hypoxia-Inducible Factors. J Mol Biol. 2024, 436: 168352. (*Co-corresponding)
4. Song W, Zhuang J, Zhang N, Ren X, Xu W, Guo M, Diao X, Liu C, Jin J, Wu D*, Zhang Y*. SAR study of 1,2-benzisothiazole dioxide compounds that agonize HIF-2 stabilization and EPO production. Bioorg Med Chem. 2023, 77: 117041. (*Co-corresponding)
5. Sun X, Jing L, Li F, Zhang M, Diao X, Zhuang J, Rastinejad F, Wu D. Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family. Proc Natl Acad Sci USA. 2022, 119: e2208804119. (Corresponding)
6. Ren X, Diao X, Zhuang J*, Wu D*. Structural basis for the allosteric inhibition of hypoxia-inducible factor 2 by belzutifan. Mol Pharmacol. 2022, 102: 240-247. (*Co-corresponding)
7. Diao X, Ye F, Zhang M, Ren X, Tian X, Lu J, Sun X, Hou Z, Chen X, Li F, Zhuang J, Ding H, Peng C, Rastinejad F*, Luo C*, Wu D*. Identification of oleoylethanolamide as an endogenous ligand for HIF-3α. Nat Commun. 2022, 13: 2529. (*Co-corresponding)
8. Zhuang J, Liu Q, Wu D*, Tie L*. Current strategies and progress for targeting the "undruggable" transcription factors. Acta Pharmacol Sin. 2022, 43: 2474-2481. (*Co-corresponding)
9. Li F, Song C, Zhang Y, Wu D. Structural overview and perspectives of the nuclear receptors, a major family as the direct targets for small-molecule drugs. Acta Biochim Biophys Sin (Shanghai). 2022, 54: 12-24. (Corresponding)
10. Wu D*, Su X, Lu J, Li S, Hood BL, Vasile S, Potluri N, Diao X, Kim Y, Khorasanizadeh S, Rastinejad F*. Bidirectional modulation of HIF-2 activity through chemical ligands. Nat Chem Biol. 2019, 15: 367-376. (*Co-corresponding) [Recommended by F1000Prime]
11. Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015, 524: 303-308. [Recommended by F1000Prime] [Highlighted in the “News&Views” section of Nature]
B. Structural & functional studies and ligand discovery for other drug target proteins
1. Li F*, Zhang M, Liu C, Cheng J, Yang Y, Peng X, Li Z, Cai W, Yu H, Wu J, Guo Y, Geng H, Fa Y, Zhang Y, Wu D*, Yin Y*. De novo discovery of a molecular glue-like macrocyclic peptide that induces MCL1 homodimerization. Proc Natl Acad Sci USA. 2025, 122: e2426006122. (*Co-corresponding)
2. Liu J, Sun X, Zhuang J, Liu Z, Xu C, Wu D*, Wu C*. Biphenyl-dihydrothiazole-cyclized peptide libraries for peptide ligand and drug discovery. Sci. China Chem. 2025, 68, 1434-1444. (*Co-corresponding)
3. Cui J, Liu X, Shang Q, Sun S, Chen S, Dong J, Zhu Y, Liu L, Xia Y, Wang Y, Xiang L, Fan B, Zhan J, Zhou Y, Chen P, Zhao R, Liu X, Xing N*, Wu D*, Shi B*, Zou Y*. Deubiquitination of CDC6 by OTUD6A promotes tumour progression and chemoresistance. Mol Cancer. 2024, 23: 86. (*Co-corresponding)
4. Li F*, Liu J, Liu C, Liu Z, Peng X, Huang Y, Chen X, Sun X, Wang S, Chen W, Xiong D, Diao X, Wang S, Zhuang J, Wu C*,Wu D*. Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-XL by engaging a single-residue discrepancy. Nat Commun. 2024, 15: 1476. (*Co-corresponding)
5. Xu M, Xu HH, Lin Y, Sun X, Wang LJ, Fang ZP, Su XH, Liang XJ, Hu Y, Liu ZM, Cheng Y, Wei Y, Li J, Li L, Liu HJ, Cheng Z, Tang N, Peng C, Li T, Liu T, Qiao L, Wu D, Ding YQ, Zhou WJ. LECT2, a ligand for Tie1, plays a crucial role in liver fibrogenesis. Cell. 2019, 178: 1478-1492. [Recommended by F1000Prime]
C. Elucidation of structures and catalytic mechanisms of biosynthetic enzymes
1. Chen Y, Jing L, Peng M, Cai C, Shi J, Ge W, Liu Y, Shang Z, Ma J*, Wu D*, Ju J*. Enzymatic insights into the unusual formation of benzolactone and benzopyran in the biosynthesis of spiromarmycin. ACS Catal. 2025, 15: 2809-2821. (*Co-corresponding)
2. Chen X, Zhao H, Wang C, Hamed M, Shang Q, Yang Y, Diao X, Sun X, Hu W, Jiang X, Zhang Y, Hirsch AKH, Wu D*, Zhuang J*. Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes. Int J Antimicrob Agents. 2024, 63: 107160. (*Co-corresponding)
3. Wang X, Chen X, Wang ZJ, Zhuang M, Zhong L, Fu C, Garcia R, Müller R, Zhang Y, Yan J*, Wu D*, Huo L*. Discovery and characterization of a myxobacterial lanthipeptide with unique biosynthetic features and anti-inflammatory activity. J Am Chem Soc. 2023, 145: 16924-16937. (*Co-corresponding)
4. Zhong L, Diao X, Zhang N, Li F, Zhou H, Chen H, Bai X, Ren X, Zhang Y*, Wu D*, Bian X*. Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis. Nat Commun. 2021, 12: 296. (*Co-corresponding) [Selected as a Featured Article in the Field of Organic Chemistry and Chemical Biology]
Link to the ResearchGate profile
https://www.researchgate.net/lab/Dalei-Wu-Lab--Protein-Structure-and-Drug-Discovery-Dalei-Wu
